Base R split()
My prior post on tidyverse split-apply-combined ended with me favouring split + map_dfr as a replacement for group_by + do. In this post I benchmark the runtime of base R split().
Looking at the runtime graphs which follow, it seems:
- Base R
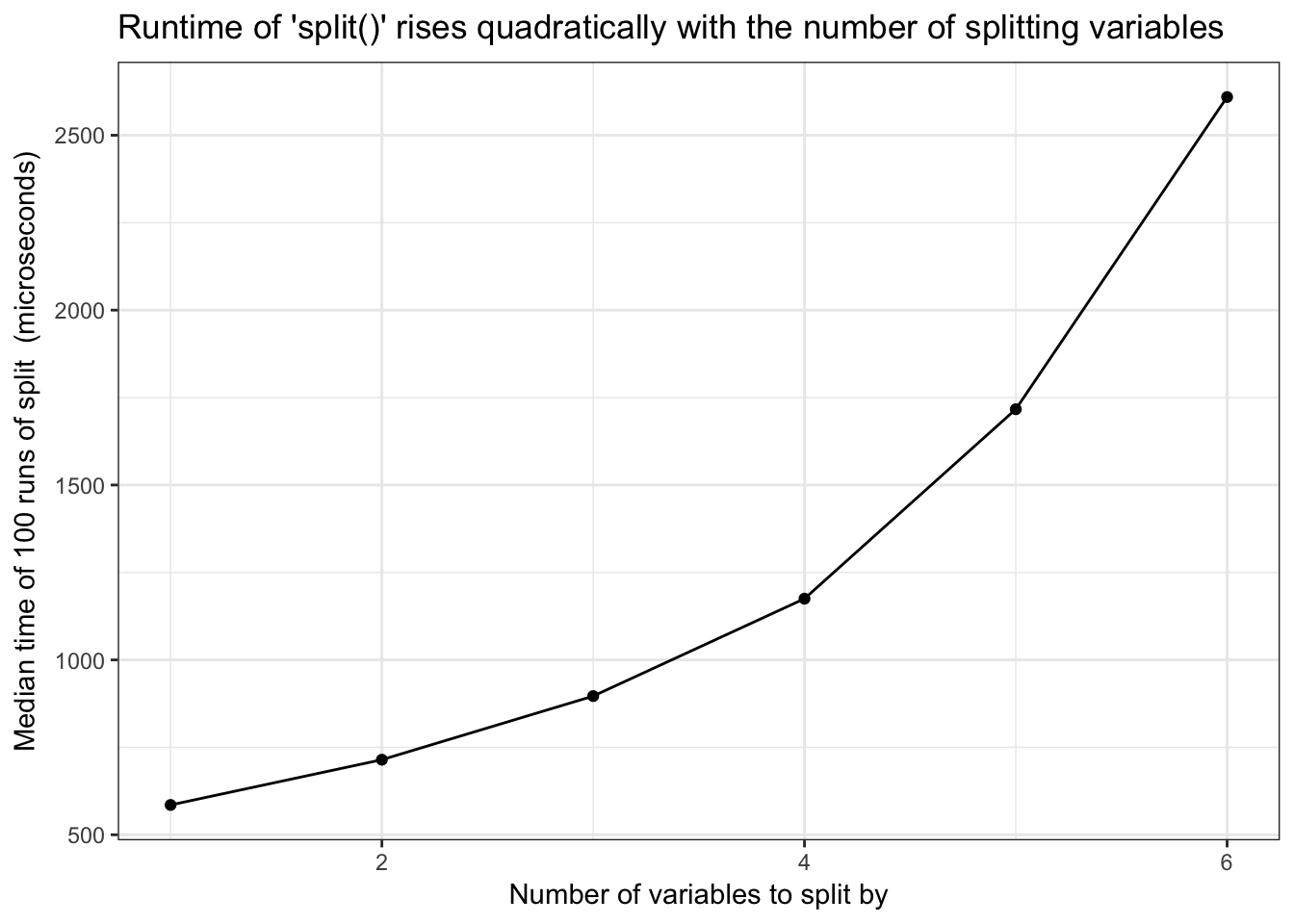
split()runtime is quadratic in number of splitting variables - Base R
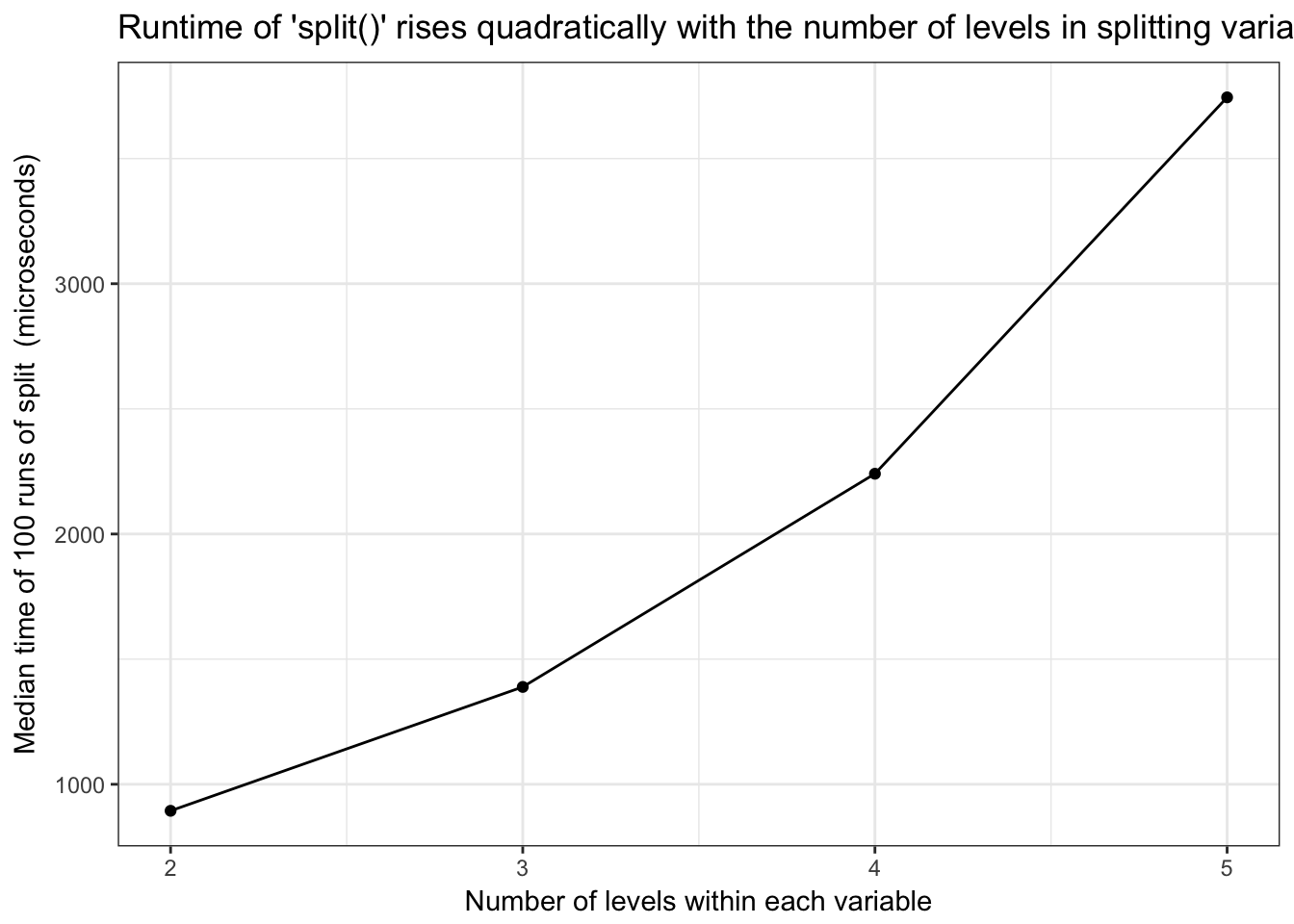
split()runtime is quadratic in number of groups within each variable
Neither of these is a good thing – a doubling in the number of splitting variables, or a doubling of levels within a splitting variable would increase runtimes by ~4x (modulo the baseline runtime)
Base R split() runtime is quadratic in number of splitting variables
test_df <- create_test_df(cols=10, rows=10, levels_per_var=2) # See appendix for 'create_test_df'
bench <- microbenchmark(
split(test_df, test_df[, c('a' )]),
split(test_df, test_df[, c('a', 'b' )]),
split(test_df, test_df[, c('a', 'b', 'c' )]),
split(test_df, test_df[, c('a', 'b', 'c', 'd' )]),
split(test_df, test_df[, c('a', 'b', 'c', 'd', 'e' )]),
split(test_df, test_df[, c('a', 'b', 'c', 'd', 'e', 'f')])
)
Base R split() runtime is quadratic in number of groups within each variable
test_df2 <- create_test_df(cols=4, rows=40, levels_per_var= 2) # See appendix for 'create_test_df'
test_df3 <- create_test_df(cols=4, rows=40, levels_per_var= 3) # See appendix for 'create_test_df'
test_df4 <- create_test_df(cols=4, rows=40, levels_per_var= 4) # See appendix for 'create_test_df'
test_df5 <- create_test_df(cols=4, rows=40, levels_per_var= 5) # See appendix for 'create_test_df'
bench <- microbenchmark(
split(test_df2 , test_df2[, c('a', 'b', 'c')]),
split(test_df3 , test_df3[, c('a', 'b', 'c')]),
split(test_df4 , test_df4[, c('a', 'b', 'c')]),
split(test_df5 , test_df5[, c('a', 'b', 'c')])
)
Appendix
set.seed(1)
#-----------------------------------------------------------------------------
#' Create a test data set
#'
#' @param cols how many columns?
#' @param rows how many rows?
#' @param levels_per_col how many distinct levels for each variable/column?
#'
#-----------------------------------------------------------------------------
create_test_df <- function(cols, rows, levels_per_var) {
data_source <- letters[seq(levels_per_var)]
create_column <- function(...) {sample(data_source, size = rows, replace = TRUE)}
letters[seq(cols)] %>%
set_names(letters[seq(cols)]) %>%
purrr::map_dfc(create_column)
}
Share this post
Twitter
Google+
Facebook
Reddit
LinkedIn
StumbleUpon
Pinterest
Email